Whirlwind tour#
Doing science in Python is fun. But it can also be a pain. Sciris can’t make it any more fun, but hopefully it can make it less of a pain. This tutorial showcases some of Sciris’ most commonly used features, which are intended to help Python get out of the way of your science. It won’t write your code for you like ChatGPT, but it does mean you have less code to write.
Click here to open an interactive version of this notebook.
Simple data operations#
While Sciris does add some new features (we’ll get to those later), its main purpose is to let you do the things you’re already doing more easily. For example, finding values in an array:
[1]:
import numpy as np
import sciris as sc
data = np.random.rand(50)
inds = sc.findinds(data>0.9)
print(f'While the mean of the data was {sc.arraymean(data)}, '
f'there were {len(inds)} values over 0.9: '
f'these were {sc.strjoin(inds)}.')
While the mean of the data was 0.55 ± 0.55, there were 8 values over 0.9: these were 2, 8, 9, 19, 25, 27, 37, 41.
I’m sure you already knew how to find indices of an array, calculate the mean and standard deviation, and turn a list of values into a string. But it’s nice if those things can be made easier, right?
Simple data containers and plotting#
Matplotlib, NumPy, and pandas are all fantastic – but often they provide lower-level interfaces than what is commonly needed, meaning that some tasks can be made even simpler.
[2]:
import pylab as pl # Equivalent to "import matplotlib.pyplot as plt", but easier to type!
# Create some data
dates = sc.daterange('2022-01-01', '2022-02-28', as_date=True) # Create a list of dates
values = 1e6*np.random.randn(31+28)**2 # Generate some values
outliers = values > 2*values.mean() # Find outliers
# Plot
data = sc.dataframe(x=dates, y=values, outliers=outliers) # Shortcut to pd.DataFrame
pl.scatter(data.x, data.y, c=data.outliers) # Vanilla Matplotlib!
sc.dateformatter() # Format a date axis nicely
sc.SIticks() # Convert the y-axis to use SI notation
pl.show()
# Describe results
mean = sc.sigfig(data.y.mean(), 3, sep=True)
print(f'The data had mean {mean} and there were {data.outliers.sum()} outliers.')
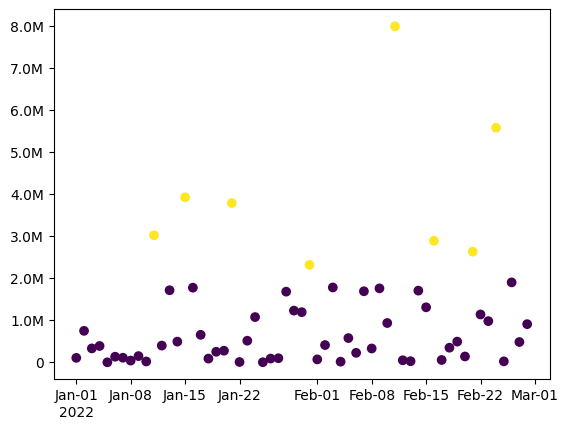
The data had mean 1,070,000 and there were 8 outliers.
Simple containers#
Can’t decide if something should be a dict or an object? Do you want the flexibility of a dict, but the convenience and explicitness of an object? Well, why not use both?
[3]:
data = sc.objdict(a=[1,2,3], b=[4,5,6])
assert data.a == data['a'] == data[0] # Flexible options for indexing
assert data[:].sum() == 21 # You can sum a dict!
for i, key, value in data.enumitems():
print(f'Item {i} is named "{key}" and has value {value}')
Item 0 is named "a" and has value [1, 2, 3]
Item 1 is named "b" and has value [4, 5, 6]
Loading and saving#
Do you have a custom object that it would be nice to be able to save and pick up where you left off?
[4]:
class Sim:
def __init__(self, days, trials):
self.days = days
self.trials = trials
def run(self):
self.x = np.arange(self.days)
self.y = np.cumsum(np.random.randn(self.days, self.trials)**3, axis=0)
def plot(self):
with pl.style.context('sciris.fancy'): # Custom plot style
pl.plot(self.x, self.y, alpha=0.6)
# Run and save
sim = Sim(days=100, trials=10)
sim.run()
sc.save('my-sim.obj', sim) # Save any Python object to disk
# Load and plot
new_sim = sc.load('my-sim.obj') # Load any Python object
new_sim.plot()
Parallelization#
Have you ever thought “Gosh, I should really parallelize this code, but it’s going to take too long, and besides doctors say you should get up and stretch everyone once in a while, so it’s OK that I’m waiting for 9 minutes out of every 10 while my code runs”? This might be for you.
[5]:
# Define the function to parallelize
def func(scale, x_offset, y_offset):
np.random.seed(scale)
data = sc.objdict()
data.scale = scale
data.x = x_offset+scale*np.random.randn(100)
data.y = y_offset+scale*np.random.randn(100)
return data
# Run in parallel
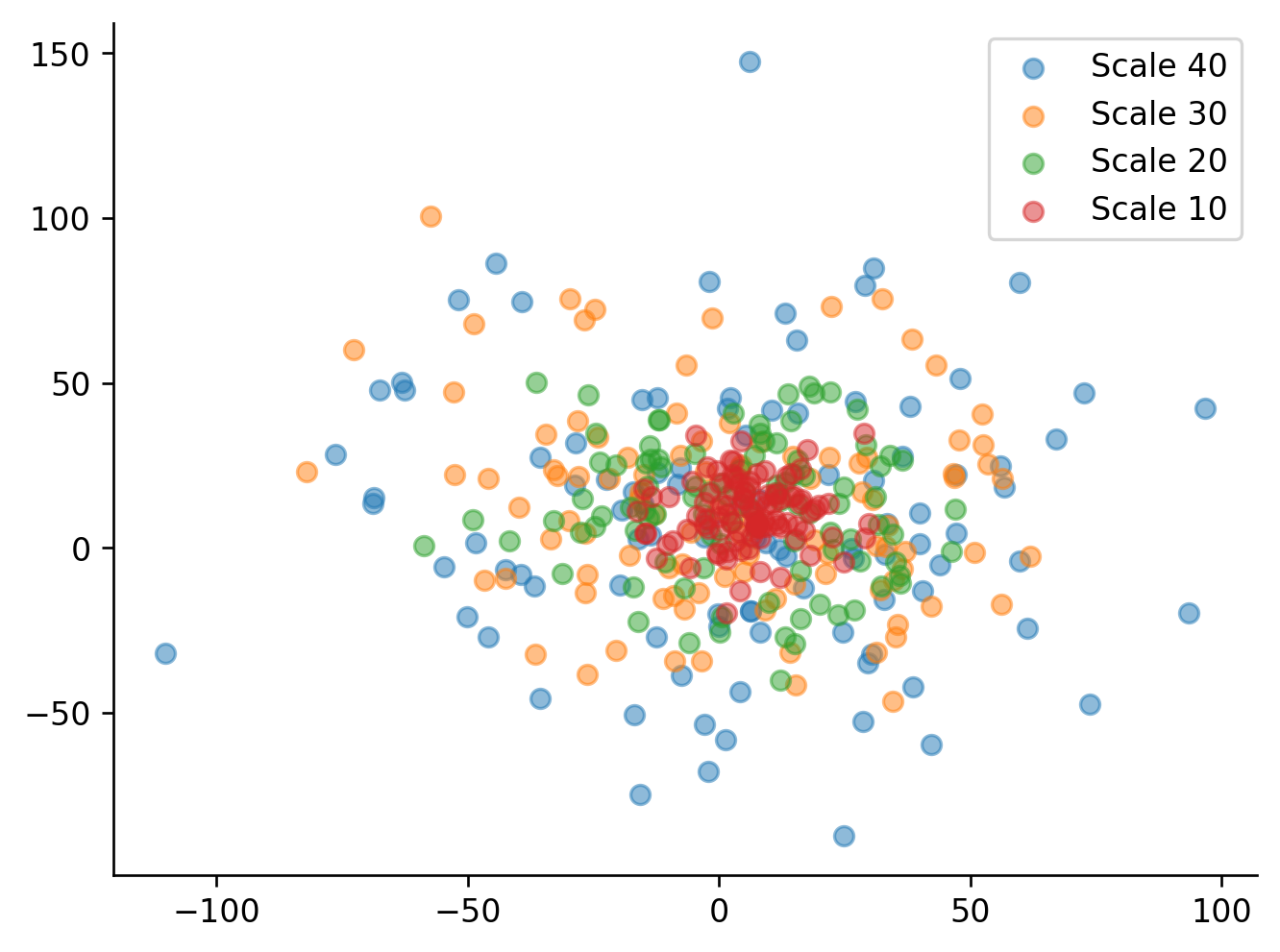
scales = [40,30,20,10] # Reverse order is easier to see when plotted
results = sc.parallelize(func, iterkwargs=dict(scale=scales), x_offset=5, y_offset=10)
# Plot
sc.options(dpi=120, jupyter=True) # Set the figure DPI and backend
for data in results:
pl.scatter(data.x, data.y, alpha=0.5, label=f'Scale {data.scale}')
pl.legend()
sc.boxoff(); # Remove top and right spines
Putting it all together#
Here’s the “showcase” example of the wave generator from the getting started page, which hopefully now makes more sense!
[6]:
# Define random wave generator
def randwave(std, xmin=0, xmax=10, npts=50):
np.random.seed(int(100*std)) # Ensure differences between runs
a = np.cos(np.linspace(xmin, xmax, npts))
b = np.random.randn(npts)
return a + b*std
# Start timing
T = sc.timer()
# Calculate output in parallel
waves = sc.parallelize(randwave, np.linspace(0, 1, 11))
# Save to files
filenames = [sc.save(f'wave{i}.obj', wave) for i,wave in enumerate(waves)]
# Create dict from files
data = sc.odict({fname:sc.load(fname) for fname in filenames})
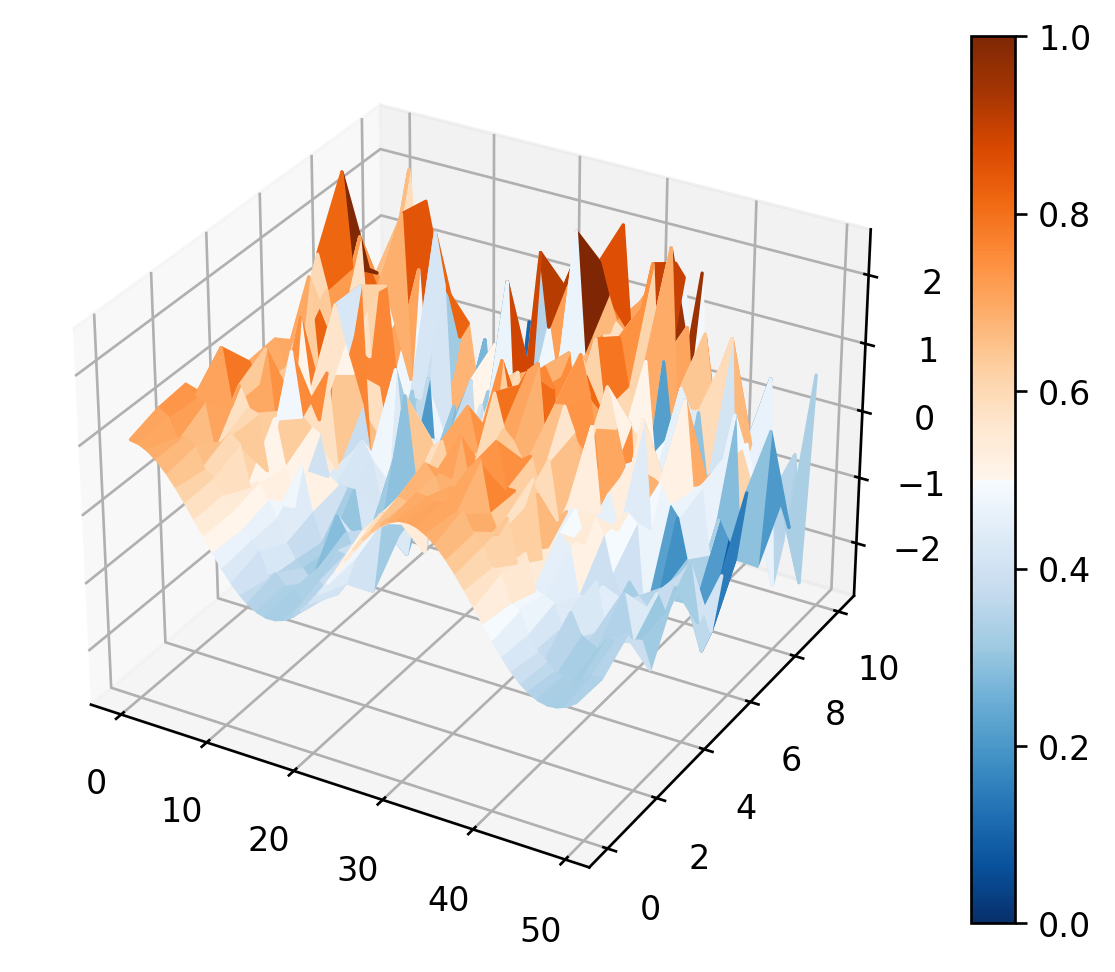
# Create 3D plot
sc.surf3d(data[:], cmap='orangeblue')
pl.show()
# Print elapsed time
T.toc('Congratulations, you finished the first tutorial')
Congratulations, you finished the first tutorial: 0.603 s
