Advanced features#
Here are yet more tools that the average user won’t need, but might come in handy one day.
Click here to open an interactive version of this notebook.
Nested dictionaries#
Nested dictionaries are a useful way of storing complex data (and in fact are more or less the basis of JSON), but can be a pain to interact with if you don’t know the structure in advance. Sciris has several functions for working with nested dictionaries. For example:
[1]:
import sciris as sc
# Create the structure
nest = {}
sc.makenested(nest, ['key1','key1.1'])
sc.makenested(nest, ['key1','key1.2'])
sc.makenested(nest, ['key1','key1.3'])
sc.makenested(nest, ['key2','key2.1','key2.1.1'])
sc.makenested(nest, ['key2','key2.2','key2.2.1'])
# Set the value for each "twig"
count = 0
for twig in sc.iternested(nest):
count += 1
sc.setnested(nest, twig, count)
# Convert to a JSON to view the structure more clearly
sc.printjson(nest)
# Get all the values from the dict
values = []
for twig in sc.iternested(nest):
values.append(sc.getnested(nest, twig))
print(f'{values = }')
{
"key1": {
"key1.1": 1,
"key1.2": 2,
"key1.3": 3
},
"key2": {
"key2.1": {
"key2.1.1": 4
},
"key2.2": {
"key2.2.1": 5
}
}
}
values = [1, 2, 3, 4, 5]
Sciris also has a sc.search() function, which can find either keys or values that match a certain pattern:
[2]:
print(sc.search(nest, 'key2.1.1'))
print(sc.search(nest, value=5))
[['key2', 'key2.1', 'key2.1.1']]
[['key2', 'key2.2', 'key2.2.1']]
There’s even an sc.iterobj() function that can make arbitrary changes to an object:
[3]:
def increment(obj):
return obj + 1000 if isinstance(obj, int) and obj !=3 else obj
sc.iterobj(nest, increment, inplace=True)
sc.printjson(nest)
{
"key1": {
"key1.1": 1001,
"key1.2": 1002,
"key1.3": 3
},
"key2": {
"key2.1": {
"key2.1.1": 1004
},
"key2.2": {
"key2.2.1": 1005
}
}
}
Context blocks#
Sciris contains two context block (i.e. “with ... as”) classes for catching what happens inside them.
sc.capture() captures all text output to a variable:
[4]:
import sciris as sc
import numpy as np
def verbose_func(n=200):
for i in range(n):
print(f'Here are 5 random numbers: {np.random.rand(5)}')
with sc.capture() as text:
verbose_func()
lines = text.splitlines()
target = '777'
for l,line in enumerate(lines):
if target in line:
print(f'Found target {target} on line {l}: {line}')
Found target 777 on line 37: Here are 5 random numbers: [0.23091441 0.92720101 0.64386704 0.98108182 0.77739743]
Found target 777 on line 40: Here are 5 random numbers: [0.10017502 0.85447773 0.70585521 0.79761481 0.56303318]
Found target 777 on line 42: Here are 5 random numbers: [0.98338691 0.23776133 0.79377702 0.38521457 0.13102052]
Found target 777 on line 57: Here are 5 random numbers: [0.48547204 0.0794115 0.83758695 0.59309727 0.48170777]
Found target 777 on line 128: Here are 5 random numbers: [0.91227649 0.17771961 0.64710676 0.36414018 0.0829745 ]
The other function, sc.tryexcept(), is a more compact way of writing try ... except blocks, and gives detailed control of error handling:
[5]:
def fickle_func(n=1):
for i in range(n):
rnd = np.random.rand()
if rnd < 0.005:
raise ValueError(f'Value {rnd:n} too small')
elif rnd > 0.99:
raise RuntimeError(f'Value {rnd:n} too big')
sc.heading('Simple usage, exit gracefully at first exception')
with sc.tryexcept():
fickle_func(n=1000)
sc.heading('Store all history')
tryexc = None
for i in range(1000):
with sc.tryexcept(history=tryexc, verbose=False) as tryexc:
fickle_func()
tryexc.disp()
————————————————————————————————————————————————
Simple usage, exit gracefully at first exception
————————————————————————————————————————————————
<class 'RuntimeError'> Value 0.995604 too big
—————————————————
Store all history
—————————————————
type value traceback
0 <class 'RuntimeError'> Value 0.992427 too big <traceback object at 0x7f3430157a80>
1 <class 'ValueError'> Value 0.0035279 too small <traceback object at 0x7f3428df4a80>
2 <class 'RuntimeError'> Value 0.991026 too big <traceback object at 0x7f3402fa3a80>
3 <class 'RuntimeError'> Value 0.996673 too big <traceback object at 0x7f3402eb74c0>
4 <class 'ValueError'> Value 0.000397334 too small <traceback object at 0x7f3402eb7400>
5 <class 'RuntimeError'> Value 0.992735 too big <traceback object at 0x7f3402eb7140>
6 <class 'RuntimeError'> Value 0.995062 too big <traceback object at 0x7f3402eb76c0>
7 <class 'RuntimeError'> Value 0.992525 too big <traceback object at 0x7f3402eb7540>
8 <class 'RuntimeError'> Value 0.993638 too big <traceback object at 0x7f3402eb6f00>
9 <class 'ValueError'> Value 0.001585 too small <traceback object at 0x7f3402eb7800>
10 <class 'ValueError'> Value 0.00207436 too small <traceback object at 0x7f3402eb7300>
11 <class 'RuntimeError'> Value 0.996381 too big <traceback object at 0x7f3402eb7880>
Interpolation and optimization#
Sciris includes two algorithms that complement their SciPy relatives: interpolation and optimization.
Interpolation#
The function sc.smoothinterp() smoothly interpolates between points but does not use spline interpolation; this makes it somewhat of a balance between numpy.interp() (which only interpolates linearly) and scipy.interpolate.interp1d(..., method='cubic'), which takes considerable liberties between data points:
[6]:
import sciris as sc
import numpy as np
import pylab as pl
from scipy import interpolate
# Create the data
origy = np.array([0, 0.2, 0.1, 0.9, 0.7, 0.8, 0.95, 1])
origx = np.linspace(0, 1, len(origy))
newx = np.linspace(0, 1)
# Create the interpolations
sc_y = sc.smoothinterp(newx, origx, origy, smoothness=5)
np_y = np.interp(newx, origx, origy)
si_y = interpolate.interp1d(origx, origy, 'cubic')(newx)
# Plot
kw = dict(lw=2, alpha=0.7)
pl.plot(newx, np_y, '--', label='NumPy', **kw)
pl.plot(newx, si_y, ':', label='SciPy', **kw)
pl.plot(newx, sc_y, '-', label='Sciris', **kw)
pl.scatter(origx, origy, s=50, c='k', label='Data')
pl.legend();
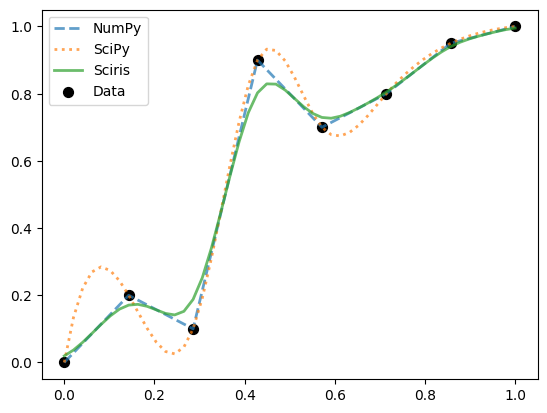
As you can see, sc.smoothinterp() gives a more “reasonable” approximation to the data, at the expense of not exactly passing through all the data points.
Optimization#
Sciris includes a gradient descent optimization method, adaptive stochastic descent (ASD), that can outperform SciPy’s built-in optimization methods (such as simplex) for certain types of optimization problem. For example:
[7]:
# Basic usage
import numpy as np
import sciris as sc
from scipy import optimize
# Very simple optimization problem -- set all numbers to 0
func = np.linalg.norm
x = [1, 2, 3]
with sc.timer('scipy.optimize()'):
opt_scipy = optimize.minimize(func, x)
with sc.timer('sciris.asd()'):
opt_sciris = sc.asd(func, x, verbose=False)
print(f'Scipy result: {func(opt_scipy.x)}')
print(f'Sciris result: {func(opt_sciris.x)}')
scipy.optimize(): 21.5 ms
sciris.asd(): 5.48 ms
Scipy result: 4.829290232718364e-08
Sciris result: 2.9796874070320123e-16
Compared to SciPy’s simplex algorithm, Sciris’ ASD algorithm was ≈3 times faster and found a result ≈8 orders of magnitude smaller.
Animation#
And finally, let’s end on something fun. Sciris has an sc.animation() class with lots of options, but you can also just make a quick movie from a series of plots. For example, let’s make some lines dance:
[8]:
pl.figure()
frames = [pl.plot(pl.cumsum(pl.randn(100))) for i in range(20)] # Create frames
sc.savemovie(frames, 'dancing_lines.gif'); # Save movie as a gif
Saving 20 frames at 10.0 fps and 150 dpi to "dancing_lines.gif" using imagemagick...
Done; movie saved to "dancing_lines.gif"
File size: 127 KB
Elapsed time: 5.50 s
This creates the following movie, which is a rather delightful way to end:
We hope you enjoyed this series of tutorials! Remember, write to us if you want to get in touch.
